Abstract
Introduction - Electronic health care data offers the opportunity to study rare events. However, detecting such outcomes in large datasets remains a large problem, as not all necessary information is recorded. The aim of this study was to develop and validate a model to identify leukemia patients with major hemorrhages (WHO grade 3 or 4) from routinely recorded and electronic search accessible clinical data.
Methods - The model was developed using routinely recorded clinical data of a cohort of leukemia patients admitted to the Leiden University Medical Center in the Netherlands between June 2011 and December 2015. Recorded variables were age, gender, DBC codes, dates of hospitalizations, received blood products, hemoglobin measurements, and dates of CT-scans of the brain. Drop in hemoglobin per 24 hours was categorized into ≤0.8, >0.8 up to and including 1.6g/dl, >1.6 to 1.9 g/dl, >1.9 to 2.2 g/dl, >2.2 to 2.8 g/dl and >2.8 g/dl. Transfusion need was defined as total number of blood products per 24 hours, including red blood cells, platelets and plasma and categorized into ≤1, 2, 3, 4, 5, and ≥6 blood products. Information about bleeding was collected via chart review of a sample of observation days. Observation days within certain strata of the indicators were oversampled. To adjust for this, days were weighted according to the prevalence of the indicators in the complete cohort for all analyses.
The final model was predefined to include CT-brain (yes/no). For drop in hemoglobin level and transfusion need the cut off values with the best discriminating capacity, as measured by the C-statistic, were included. The model was internally validated using bootstrap resampling with 100 repetitions. Performance of the model was expressed as sensitivity, specificity, negative and positive predictive value, and the C-statistic. In addition, we calculated the number of observation days needed to screen to detect one case of major hemorrhage. External validation was performed in a cohort of leukemia patients of two other academic hospitals.
Results - The derivation cohort consisted of 255 patients comprising 10,638 observation days. Chart review was performed for 353 days. Within this sample, 19 cases of major hemorrhage were found, corresponding to 16 unique patients. Extrapolated to the complete cohort, the incidence of major hemorrhage was 0.22 per 100 observation days. The final model consisted of CT-brain (yes/no), hemoglobin drop of ≥0.8 g/dl and need of six or more transfusions. The C-statistic was 0.988 (95% confidence interval (CI) 0.981 to 0.995). Presence of at least one of the indicators had a sensitivity of 100%, a specificity of 93.1% and a positive predictive value of 3.1%.
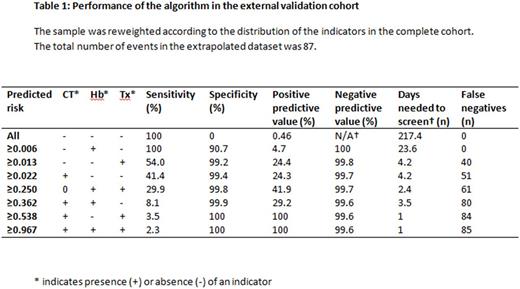
The external validation cohort consisted of 436 patients, 19,188 observation days. Chart review was performed for 599 observation days. Forty-two patients, (9.6%) experienced a major hemorrhage, corresponding to an incidence of 0.46 per 100 observation days. The C-statistic of the model was 0.975 (CI 0.970 to 0.980). Presence of at least one indicator had a sensitivity of 100%, a specificity of 90.7% and a positive predictive value of 4.7% (table 1). Without the model, the number of observation days needed to screen to detect one case of major hemorrhage was 217.4, whereas the use of this model reduces the days needed to screen to 23.6.
Conclusion - A model based on drop in hemoglobin ≥0.8g/dL, the need of ≥6 transfusions and CT-scan of the brain allows the capture of cases with major hemorrhages in large datasets over a long follow-up period while minimizing costs and effort. This model will have particular significance for researchers and blood services who aim to investigate major hemorrhage among hematological patients with sufficient sample size.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.